A function to carry diagnostics on the PCA results for a
Spectra object. Basically a wrapper to Filzmoser's
pcaDiagplot which colors everything according to
the scheme stored in the Spectra object. Works with PCA
results of either class prcomp or class princomp. Works
with either classical or robust PCA results.
pcaDiag(
spectra,
pca,
pcs = 3,
quantile = 0.975,
plot = c("OD", "SD"),
use.sym = FALSE,
...
)Arguments
- spectra
An object of S3 class
Spectra().- pca
An object of class
prcompmodified to include a character string ($method) describing the pre-processing carried out and the type of PCA performed.- pcs
As per
pcaDiagplot. The number of principal components to include.- quantile
As per
pcaDiagplot. The significance criteria to use as a cutoff.- plot
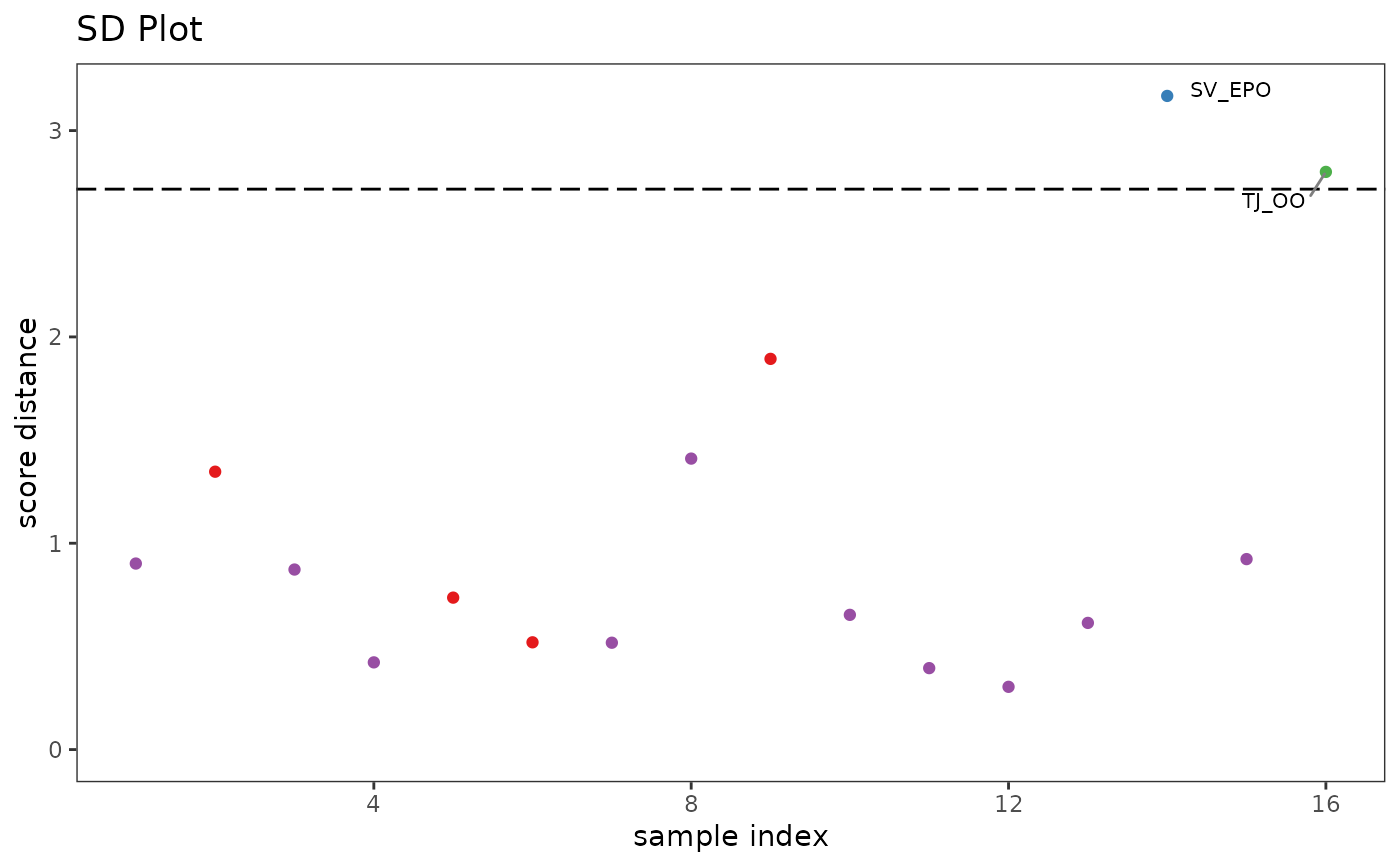
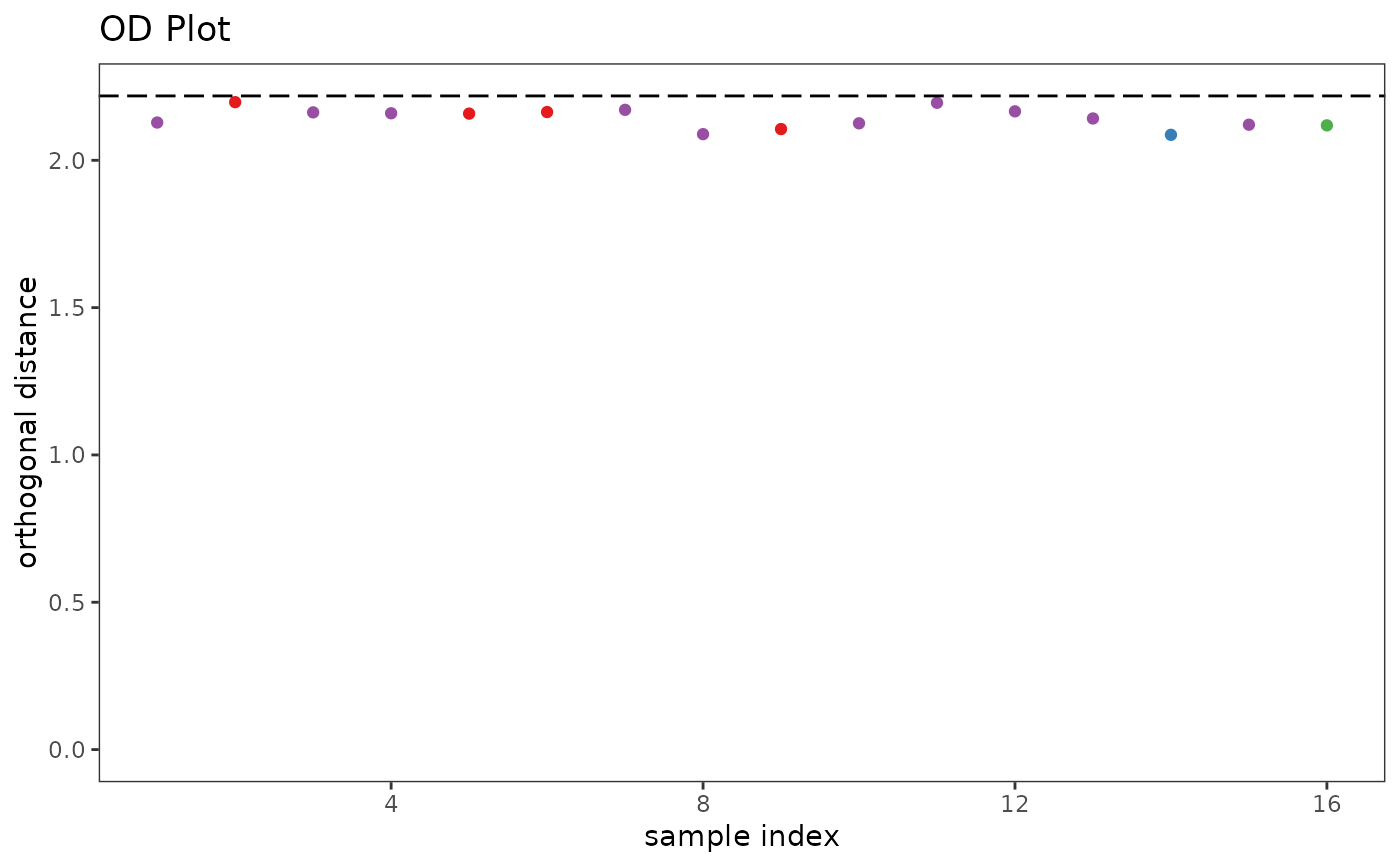
A character string, indicating whether to plot the score distances or orthogonal distances, or both. Options are
c("OD", "SD").- use.sym
logical; if true, the color scheme is change to black and symbols are used for plotting.
- ...
Parameters to be passed to the plotting routines. Applies to base graphics only.
Value
The returned value depends on the graphics option selected (see ChemoSpecUtils::GraphicsOptions()).
base A data frame or list containing the data plotted. Assign the value and run
str()ornames()on it to see what it contains. Side effect is a plot.ggplot2 The plot is displayed, and a
ggplot2plot object is returned if the value is assigned. The plot can be modified in the usualggplot2manner. If you want the plotted values, you can access them via the base graphics mode.
Details
If both plots are desired, the output should be directed to a file rather
than the screen. Otherwise, the 2nd plot overwrites the 1st in the active
graphics window. Alternatively, just call the function twice, once
specifying OD and once specifying SD.
References
K. Varmuza and P. Filzmoser Introduction to Multivariate Statistical Analysis in Chemometrics, CRC Press, 2009.
See also
pcaDiagplot in package
chemometrics for the underlying function. Additional documentation at
https://bryanhanson.github.io/ChemoSpec/
Examples
# This example assumes the graphics output is set to ggplot2 (see ?GraphicsOptions).
library("ggplot2")
data(SrE.IR)
pca <- c_pcaSpectra(SrE.IR, choice = "noscale")
p1 <- pcaDiag(SrE.IR, pca, pcs = 2, plot = "OD") + ggtitle("OD Plot")
p1
 p2 <- pcaDiag(SrE.IR, pca, pcs = 2, plot = "SD") + ggtitle("SD Plot")
p2
p2 <- pcaDiag(SrE.IR, pca, pcs = 2, plot = "SD") + ggtitle("SD Plot")
p2