Hierarchical Cluster-Based Peak Alignment on a Spectra Object
Source:R/clupaSpectra.R
clupaSpectra.RdThis function is a wrapper to several functions in the speaq package. It implements the CluPA algorithm described in the reference.
clupaSpectra(spectra, bT = NULL, ...)Arguments
- spectra
An object of S3 class
Spectra().- bT
Numeric. The baseline threshold. Defaults to five percent of the range of the data, in
spectra$data. Passed todetectSpecPeaks.- ...
Other arguments to be passed to the underlying functions.
Value
A modifed Spectra object.
References
Vu TN, Valkenborg D, Smets K, Verwaest KA, Dommisse R, Lemiere F, Verschoren A, Goethals B, Laukens K. "An integrated workflow for robust alignment and simplified quantitative analysis of NMR spectrometry data" BMC Bioinformatics vol. 12 pg. 405 (2011).
See also
Additional documentation at https://bryanhanson.github.io/ChemoSpec/
Examples
# This example assumes the graphics output is set to ggplot2 (see ?GraphicsOptions).
# You need to install package speaq for this example
if (requireNamespace("speaq", quietly = TRUE)) {
library("ggplot2")
data(alignMUD)
p1 <- plotSpectra(alignMUD,
which = 1:20, lab.pos = 4.5, offset = 0.1,
yrange = c(0, 5000), amp = 500
)
p1 <- p1 + ggtitle("Misaligned NMR Spectra") +
coord_cartesian(xlim = c(1.5, 1.8), ylim = c(0, 1900))
p1
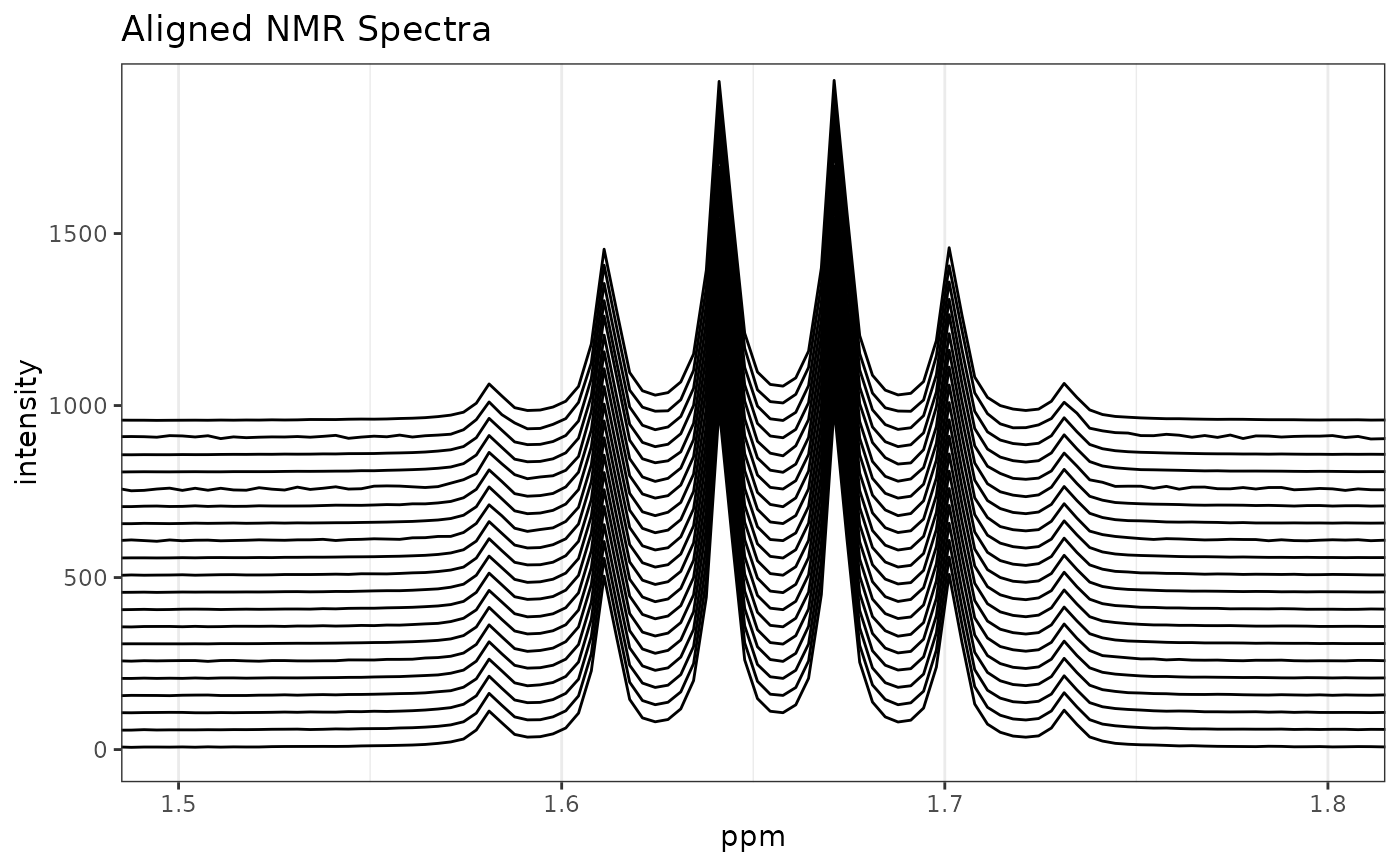
aMUD <- clupaSpectra(alignMUD)
p2 <- plotSpectra(aMUD,
which = 1:20, lab.pos = 4.5, offset = 0.1,
yrange = c(0, 5000), amp = 500
)
p2 <- p2 + ggtitle("Aligned NMR Spectra") +
coord_cartesian(xlim = c(1.5, 1.8), ylim = c(0, 1900))
p2
}
#> Coordinate system already present. Adding new coordinate system, which will
#> replace the existing one.
#>
#> Spectrum 1 has 4 peaks
#> Spectrum 2 has 4 peaks
#> Spectrum 3 has 4 peaks
#> Spectrum 4 has 4 peaks
#> Spectrum 5 has 4 peaks
#> Spectrum 6 has 4 peaks
#> Spectrum 7 has 4 peaks
#> Spectrum 8 has 4 peaks
#> Spectrum 9 has 4 peaks
#> Spectrum 10 has 4 peaks
#> Spectrum 11 has 4 peaks
#> Spectrum 12 has 4 peaks
#> Spectrum 13 has 4 peaks
#> Spectrum 14 has 4 peaks
#> Spectrum 15 has 4 peaks
#> Spectrum 16 has 4 peaks
#> Spectrum 17 has 4 peaks
#> Spectrum 18 has 4 peaks
#> Spectrum 19 has 4 peaks
#> Spectrum 20 has 4 peaks
#> --------------------------------
#> CluPA will run with maxShift= 100
#> If you want CluPA automatically detect the optimal maxShift,
#> let try with dohCluster(...,maxShift=NULL,..)
#> --------------------------------
#>
#> aligning spectrum 1
#> aligning spectrum 2
#> aligning spectrum 3
#> aligning spectrum 4
#> aligning spectrum 5
#> aligning spectrum 6
#> aligning spectrum 7
#> aligning spectrum 9
#> aligning spectrum 10
#> aligning spectrum 11
#> aligning spectrum 12
#> aligning spectrum 13
#> aligning spectrum 14
#> aligning spectrum 15
#> aligning spectrum 16
#> aligning spectrum 17
#> aligning spectrum 18
#> aligning spectrum 19
#> aligning spectrum 20
#> Median pearson correlation of aligned spectra: 0.9999948
#> Alignment time: 6e-04 minutes
#> Coordinate system already present. Adding new coordinate system, which will
#> replace the existing one.