Carry out PARAFAC analysis of a Spectra2D object.
Function parafac from multiway is used.
For large data sets, computational time may be long enough that
it might desirable to run in batch mode and possibly use parallel processing.
pfacSpectra2D(spectra, parallel = FALSE, setup = FALSE, nfac = 2, ...)Arguments
- spectra
An object of S3 class
Spectra2D.- parallel
Logical. Should parallel processing be used? Unless you love waiting, you should use parallel processing for larger data sets. If you are working on a shared machine and/or another process (created by you or another user) might also try to access all or some of the cores in your CPU, you should be careful to avoid hogging the cores.
parallel::detectCores()will tell you how many cores are available to everyone. You can runoptions(mc.cores = 2)to set the number of cores this function will use.- setup
Logical. If
TRUEthe parallel environment will be automatically configured for you. IfFALSE, the user must configure the environment themselves (desirable for instance if working on Azure or AWS EC2).- nfac
Integer. The number of factors/components to compute.
- ...
Additional parameters to be passed to function
parafac. You should give thought to value ofconst, allowed options can be seen inconst. The default is to compute an unconstrained solution. However, in some cases one may wish to apply a non-negativity constraint. Also, to suppress the progress bar, you can useverbose = FALSE.
Value
An object of class pfac and parafac, modified to include a list
element called $method which is parafac.
Warning
To get reproducible results you will need to set.seed(). See the example.
References
R. Bro "PARAFAC. Tutorial and applications" Chemometrics and Intelligent Laboratory Systems vol. 38 pgs. 149-171 (1997).
A. Smilde, R. Bro and P. Geladi "Multi-way Analysis: Applications in the Chemical Sciences" Wiley (2004).
See also
For other data reduction methods for Spectra2D objects, see
miaSpectra2D and popSpectra2D.
Examples
library("ggplot2")
data(MUD1)
set.seed(123)
res <- pfacSpectra2D(MUD1, parallel = FALSE, nfac = 2)
#>
|
| | 0%
|
|======= | 10%
|
|============== | 20%
|
|===================== | 30%
|
|============================ | 40%
|
|=================================== | 50%
|
|========================================== | 60%
|
|================================================= | 70%
|
|======================================================== | 80%
|
|=============================================================== | 90%
|
|======================================================================| 100%
# plotScores uses ggplot2 graphics
p1 <- plotScores(MUD1, res, leg.loc = "topright", ellipse = "cls")
#> Error in plotScores.Spectra2D(MUD1, res, leg.loc = "topright", ellipse = "cls"): object 'res' not found
p1 <- p1 + ggtitle("PARAFAC Score Plot")
#> Error: object 'p1' not found
p1
#> Error: object 'p1' not found
# plotLoadings2D uses base graphics
res1 <- plotLoadings2D(MUD1, res,
load_lvls = c(1, 5, 10, 15, 25),
main = "PARAFAC Comp. 1 Loadings")
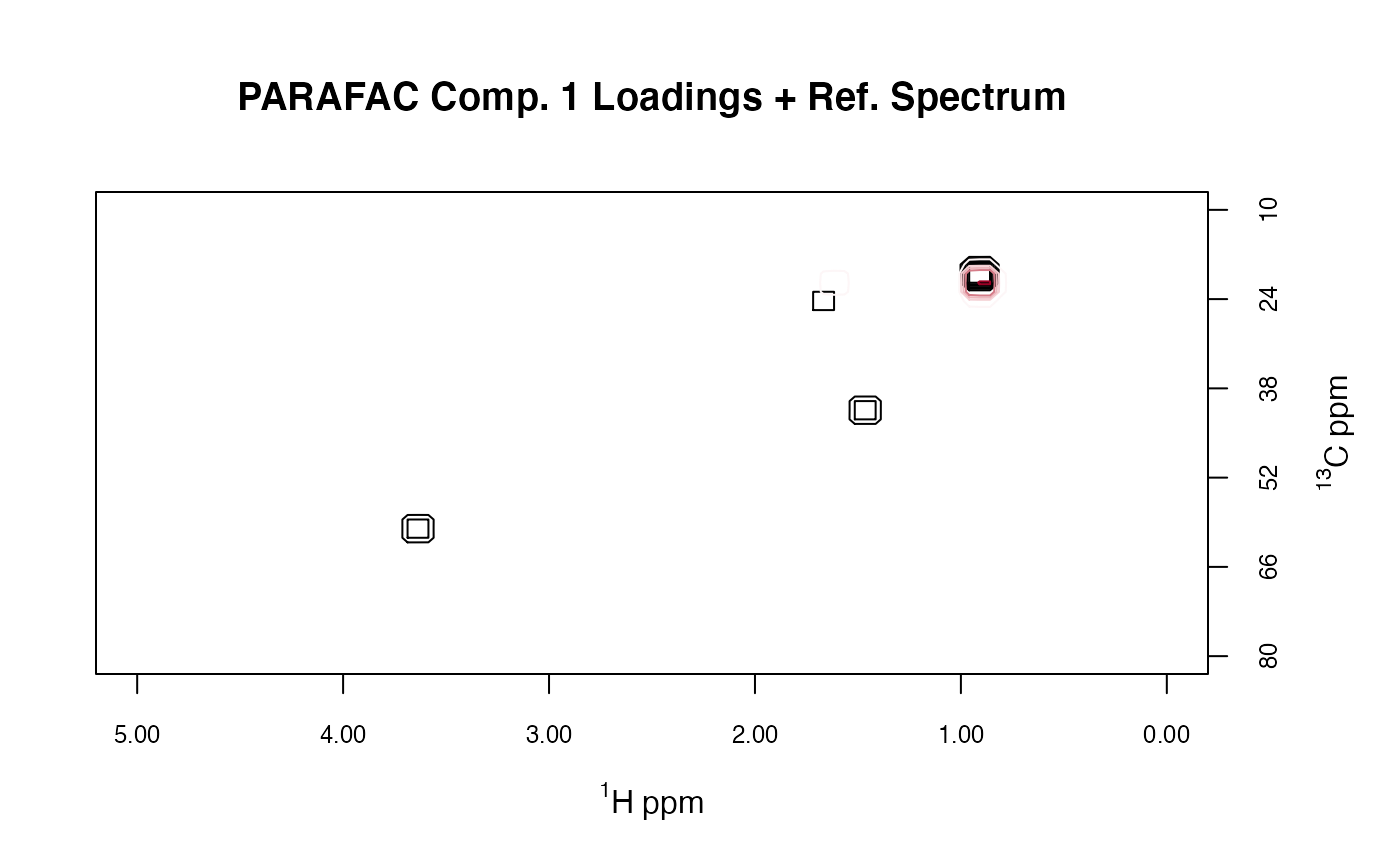
 res2 <- plotLoadings2D(MUD1, res,
load_lvls = c(1, 5, 10, 15, 25),
ref = 2, ref_lvls = seq(5, 35, 5),
ref_cols = rep("black", 7),
main = "PARAFAC Comp. 1 Loadings + Ref. Spectrum")
res2 <- plotLoadings2D(MUD1, res,
load_lvls = c(1, 5, 10, 15, 25),
ref = 2, ref_lvls = seq(5, 35, 5),
ref_cols = rep("black", 7),
main = "PARAFAC Comp. 1 Loadings + Ref. Spectrum")
 # Selection of loading matrix levels can be aided by the following
# Use res1$names to find the index of the loadings
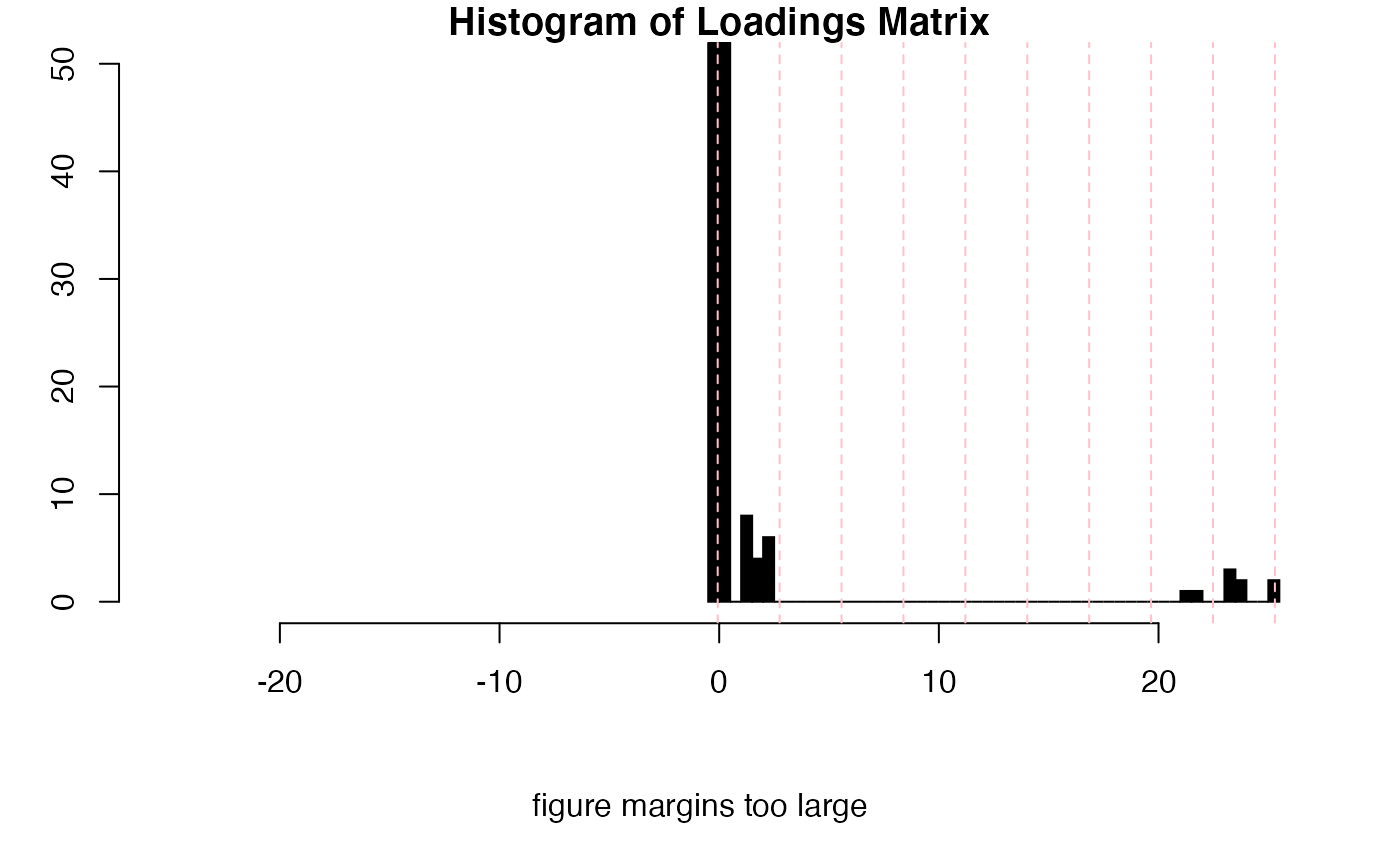
inspectLvls(res1,
which = 11, ylim = c(0, 50),
main = "Histogram of Loadings Matrix")
# Selection of loading matrix levels can be aided by the following
# Use res1$names to find the index of the loadings
inspectLvls(res1,
which = 11, ylim = c(0, 50),
main = "Histogram of Loadings Matrix")